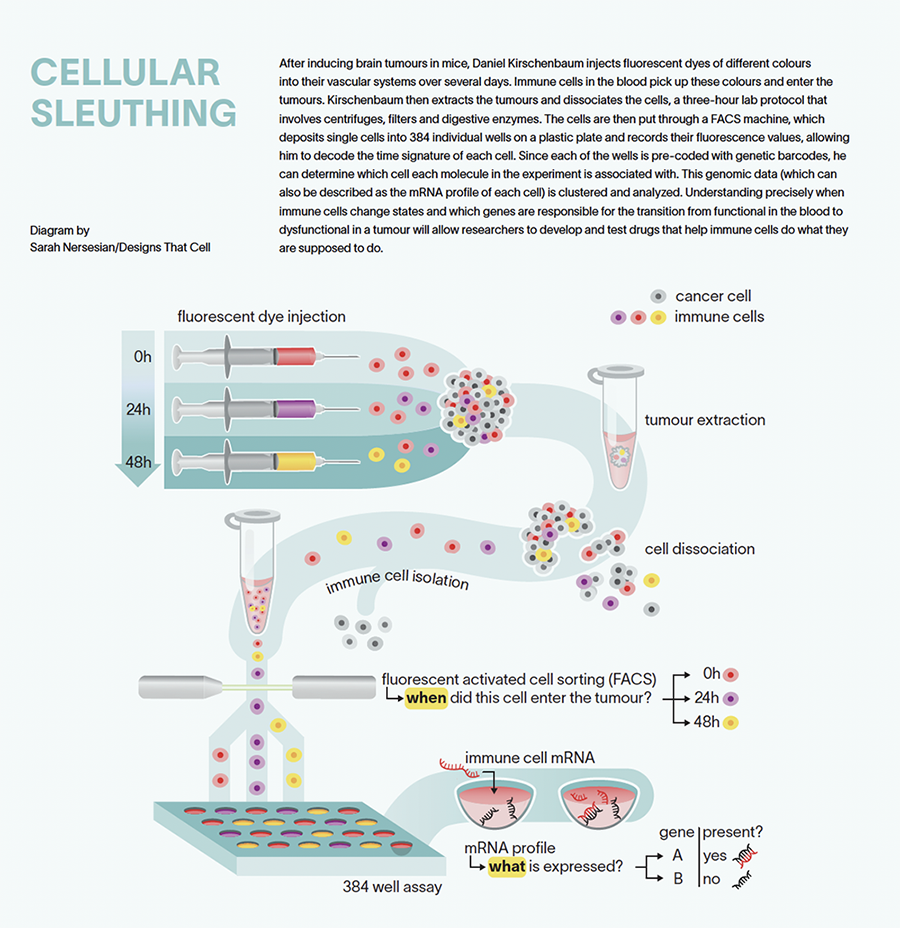
To differentiate by colour, Kirschenbaum first extracts the tumour and dissociates the cells, a three-hour lab protocol that involves centrifuges, filters and the application of digestive enzymes to disintegrate the matrix around the cells so individual cells can be isolated. The cells are then put through a FACS (fluorescence-activated cell sorting) machine, which deposits single cells into 384 individual wells on a plastic plate and records their fluorescent values, allowing him to decode the colour or time signature of each cell. Next, using enzymes that can read an RNA sequence, he amplifies the genetic material of each cell to obtain a stronger signal and puts them into a next-generation sequencer. Since each of the 384 wells is pre-coded with genetic barcodes, he can determine which cell each molecule in the experiment is associated with in addition to its time signature. This transcriptomic or genomic data (which can also be described as the mRNA profile of each cell) is clustered and analyzed by computational scientists in Amit’s lab, and Kirschenbaum and his colleagues interpret the results together.
“The fluorescent profile of the cells represents their exposure to the temporally defined injections of fluorescent dyes,” he explains. “Using this method, we know the transcriptomic profile and the temporal profile of every single cell we sorted. Based on the temporal signature we can now order the cells in time. This way we see how the transcriptomic profiles of single cells change in sequence across time.”
Parallel to this research at Weizmann, two different drugs have shown in tests the potential to modulate how immune cells interact with cancer cells, and one is close to clinical trial. But researchers don’t understand exactly how they work, says Kirschenbaum. Using the zman-seq method, “we’ll be able to see — beautifully — how certain cells respond to this drug across time,” he says. “We’ll be able to draw arrows and look at how cells change compared to the non-treated ones. We’ll be able to see and understand pathways. This is very concrete.” If an immune cell enters a tumour and a certain gene is not expressed, for example, and several hours later that gene is highly expressed, researchers will be able to discern its importance.
“It provides a fresh approach to developing better therapies,” Amit says about zman-seq’s potential to illuminate the environment in which immune cells become pathological. “Daniel’s technology to move from snapshots to movies allows us to see where the ‘bad character’ is. Now we can come up with innovative ideas for therapies that prevent that character from changing the immune cell.”
These days, while writing a paper about his method, Kirschenbaum is also trying to make it more effective. He’d rather use genetic barcodes than colours to time-stamp immune cells, because the range of useable colours is limited, whereas the number of genetic sequences is endless, plus these labels would last longer and provide more precise, higher-resolution pictures.
Zman-seq and any iterations that emerge can be used with all tumours, not just brain cancer, and this research fits with the overarching goal of developing novel immunotherapies in Amit’s lab. Yet Kirschenbaum remains somewhat restless. The work satisfies his curiosity. He finds it very stimulating and inspiring. “But at the same time,” he says, “it makes me think about next steps and new directions.”